Simulation
Purpose
1:Check the stability of hairpin DNAs at room temperature environment.
2:Determine if Hybridization Chain Reaction actually occurs
3:Whether an initiator is generated from the aptamer complex.
Evalution of DNA stability
To achieve the first purpose we use "NUPACK"(7) to determin the hairpin structure. For the following strand combination, simulation carried out using NUPACK. The length of the stem and loop was adjusted according to the reference.(8) The results are shown in Fig. 1-2.
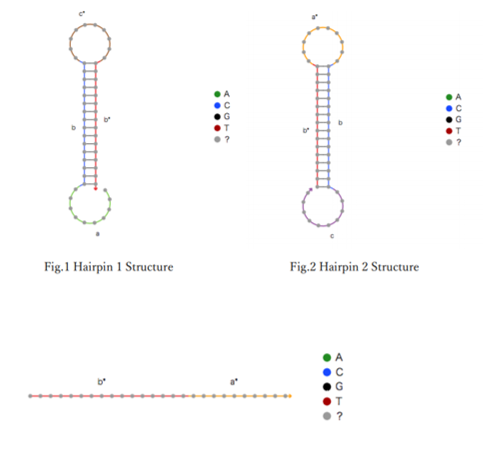
Fig.3 Initiator structure
Next, we designed the DNA sequence based on the structure of the initiator and hairpin. The sequence of the determined DNA is shown in Figure 4.
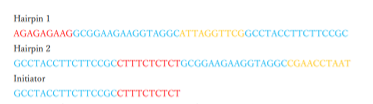
Fig.4 DNA seqence base
H1, H2, Initiator (H1, H2, Initiator = 1.0 μM).
We used NUPACK to determine the temperature stability of the three complexes (H1+H2+Initiator). The result of structural stability between 4-90˚C is shown below.
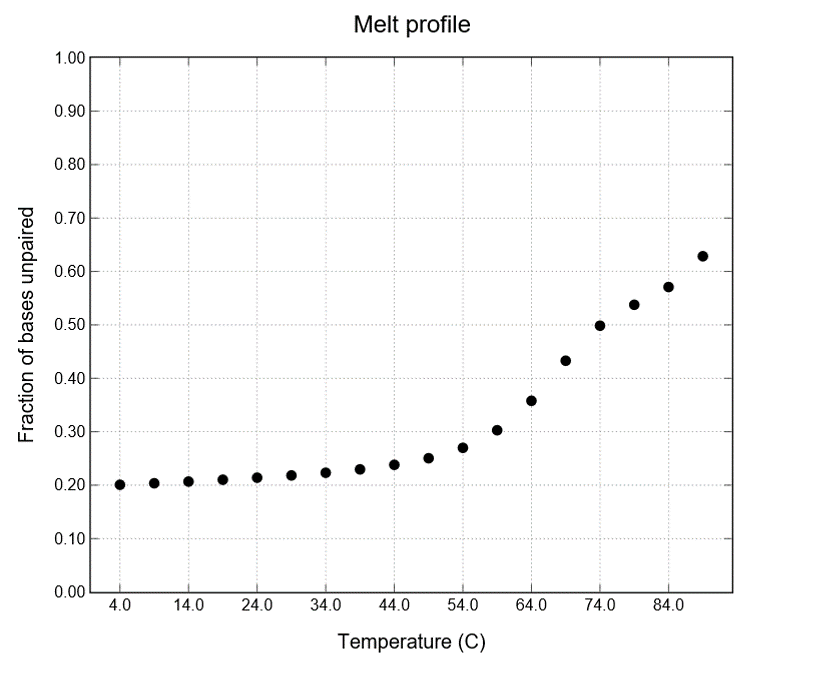
Fig.5 Melt profile of H1-H2-Initiator Structure
Figure 5 shows that the closer the structure is to red, the more stable it is. Look at the three complexes on the left of the figure 6. It shows a red color, indicating that it is stable. We found that this structure is stable at room temperature. We assume that the test kit will be used in a room temperature environment. From the results in this figure, the secondary structure should not collapse even at room temperature. So we can get accurate results. This result shows that the test kit is not affected in a room-temperature environment.
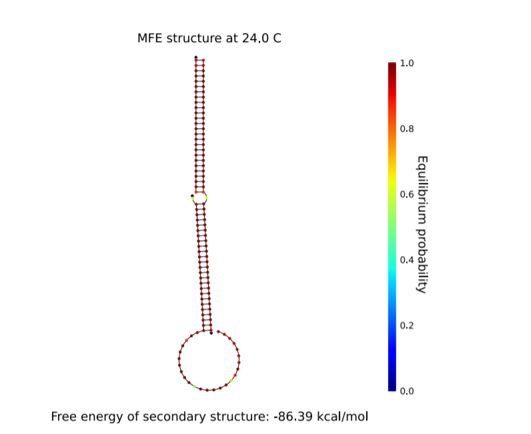
Fig.6 The stability of three complexes at 24℃
This is hairpin of H1-H2-Initiator structure at 24˚C.
Consideration of whether HCR will occur
To achieve the second purpose. we confirmed HCR simulation using Visual DSD(9). Firstly, when Inisiator was introduced in the presence of two hairpin DNAs, it was confirmed whether HCR progressed by the two hairpin DNAs. The reaction system is shown in Fig7.
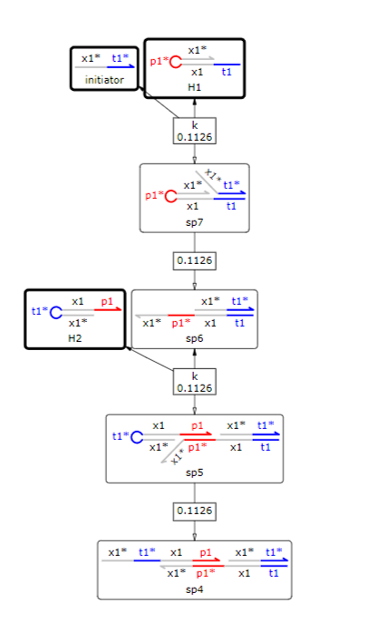
Fig.7 Hybridization reactions of initiator and hairpin DNA
The result of time-dependent concentration change of each DNA obtained from the above reaction system is shown in the Fig.8.
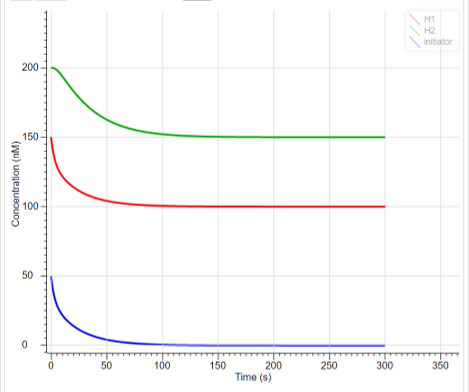
Fig.8 Changes in the concentration of reactants in HCR
We found that H1 was opened by the Initiator, then the hairpin of H2 was opened by the exposed toehold of H1 and bound in a chain reaction with it. This result suggests that the designed DNA promotes HCR properly.
In reality, the stem of the hairpin may open, causing a leak reaction in which the HCR progresses without an initiator. Therefore,we evaluated the stability of the system considering the leak reaction. The modified reaction system is shown in fig.9.
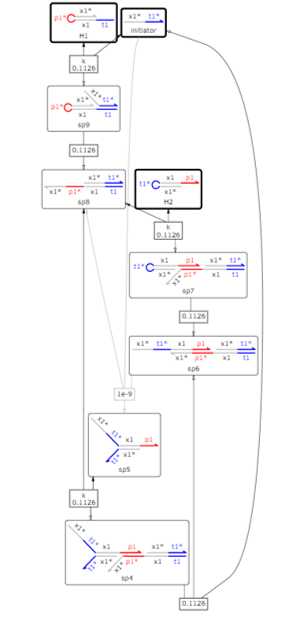
Fig.9 Reactions considered when incorporating a leak reaction in HCR
The results of time-dependent changes in the concentration of each DNA obtained from the above reaction system are shown in the figure.10.
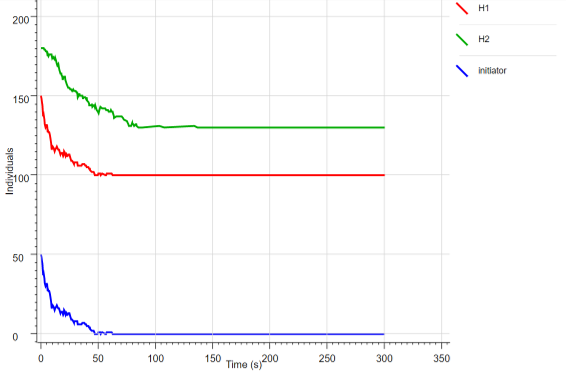
Fig.10 Changes in the concentration of each reactant in HCR when the leak reaction is introduced
By using stochastic simulation, although there is some blurring, both the initiator and hairpin tend to decrease every hour, and we expect that the reaction can actually occur.
Whether an initiator is generated from the aptamer complex.
Using Visual DSD, we investigated whether an initiator was generated when there was a sequence that binds complementarily to an aptamer.
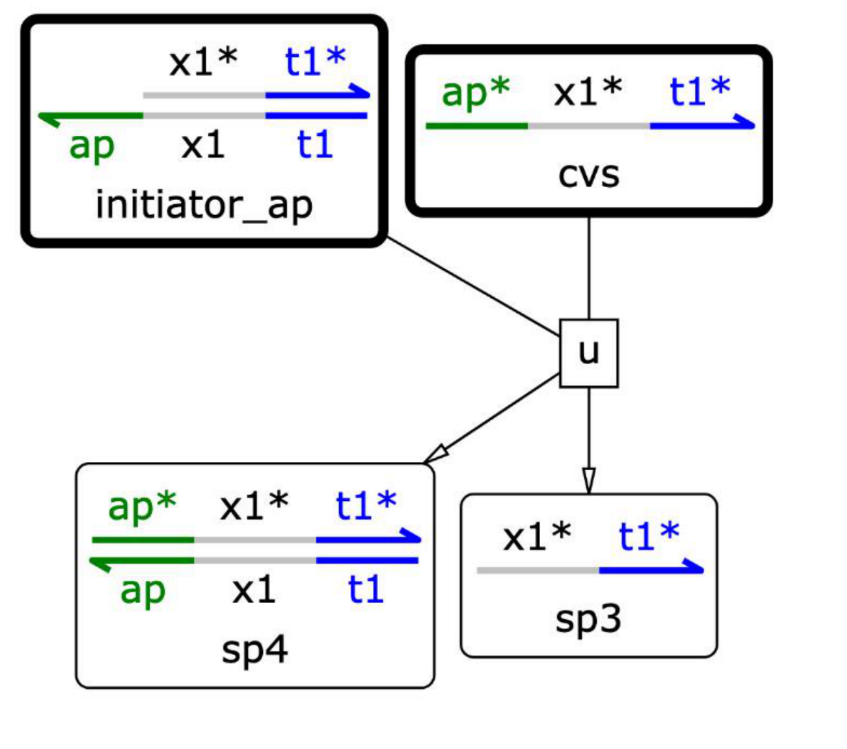
Fig.11 Reaction of aptamer-initiator complex with Angiotensin-converting enzyme 2(ACE2)
The time change of the concentration of each substance is shown in Fig.12.
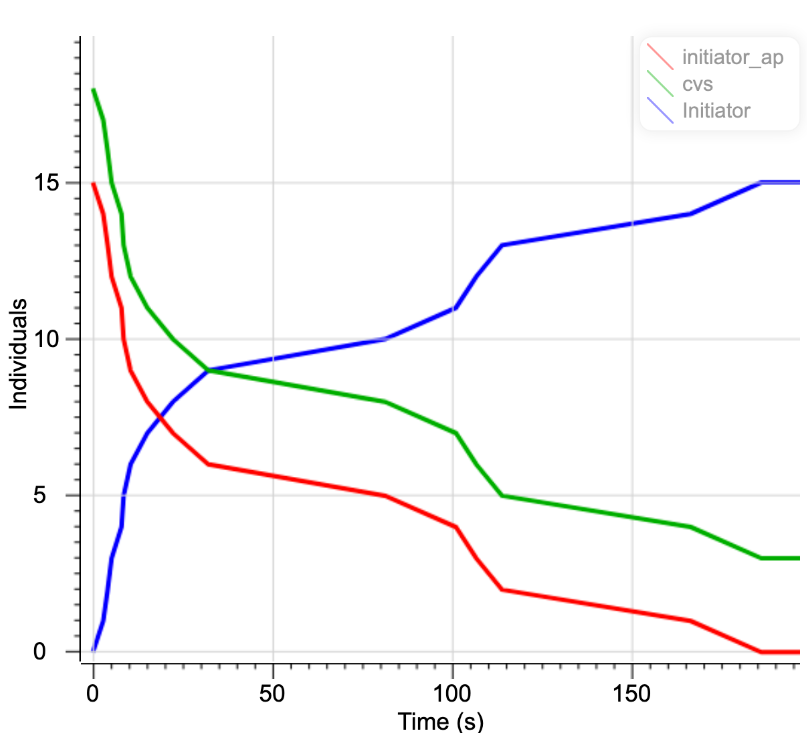
Fig.12 Changes in the concentration of reactants in the reaction of aptamer-initiator complex with ACE2(Generation of initiator)
Over time, the concentration of the aptamer-initiator complex decreased and the initiator was released from aptamer-initiator complex.
Reference
(7)http://www.nupack.org
(8)Li, Shaofei, et al. "Elucidation of leak-resistance DNA hybridization chain reaction with universality and extensibility." Nucleic Acids Research 48.5 (2020): 2220-2231.
(9)https://www.microsoft.com/en-us/research/wp-content/uploads/2016/02/dna-manual.pdf
https://classicdsd.azurewebsites.net/